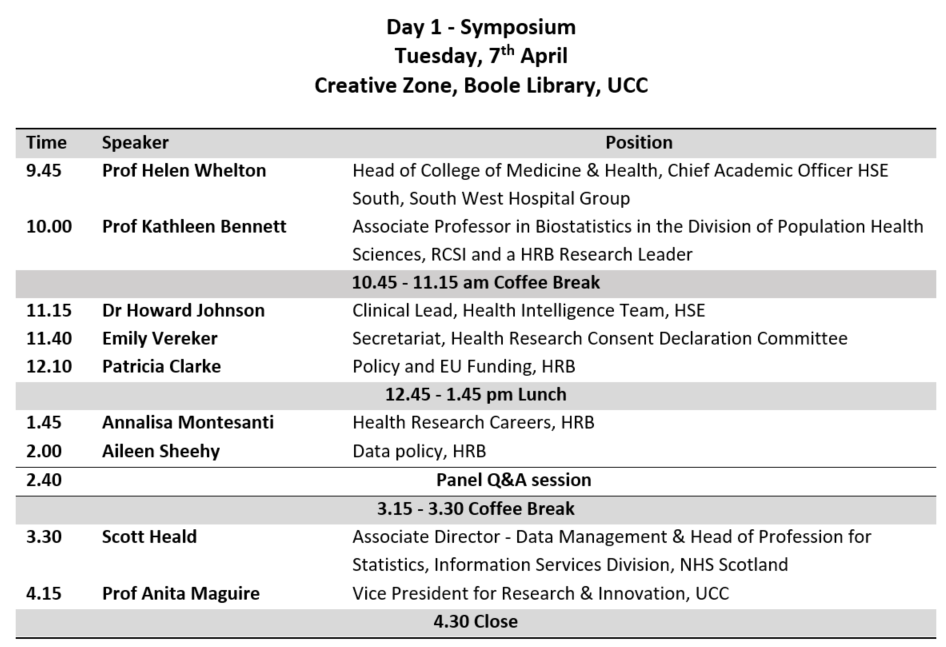
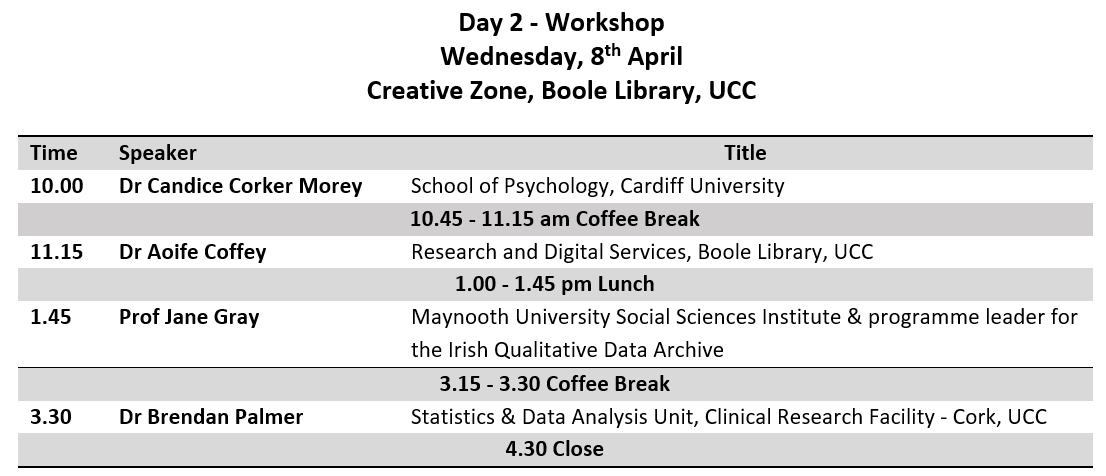
This is a tutorial on how to use R to evaluate a previously published prediction tool in a new dataset. Most of the good ideas came from Maarten van Smeden, and any mistakes are surely mine. This post is not intended to explain they why one might do what follows, but rather how to do it in R.
It is based on a recent analysis we published (in press) that validated the HOMR model to predict all-cause mortality within one-year of a hospitalization in a cohort of patients aged 65 years or older that were under the care of geriatric medicine service at Cork University Hospital (2013-01-01 to 2015-03-06). The predictors used in the HOMR model were generally things that are easy enough to collect during hospitalization. You can read all about it in the paper linked above.
All materials for the analysis can be found on the project’s OSF page.
data <- read_csv("https://crfcsdau.github.io/public/homr_data.csv")
data$alive <- factor(data$alive, levels = c("Yes", "No"))
Calculating the HOMR score
The first step in evaluating the HOMR model in our cohort of patients was to calculate the linear predictor (Z) for each of them. This was done by multiplying the fitted regression coefficients (on the log-odds scale; found in appendix E of the HOMR development paper) by each patient’s respective variable values, and then adding them all together, including the intercept term (data$z_homr).
The original HOMR development paper also provided a set of tables to calculate a score based on the predictors (data$homr_3). However, these scores are just a rescaled version of Z that gives you a set of integer values that can be easily added together from simple tables to get a prediction. However, since there was considerable rounding error (as you can see from the plot below), we used Z instead of the HOMR score in our evaluation.
Figure 1. The relationship between the HOMR score and the HOMR linear predictor (Z) in our cohort of patients.
ggplot(data, aes(x = z_homr, y = homr_3)) +
geom_point(alpha = 0.2, size = 2, color = viridis(1, begin = 0.5)) +
xlab("HOMR linear predictor (log odds scale)") +
ylab("HOMR score") +
theme_minimal()

Evaluating the model: Overview
To evaluate the HOMR Model, we followed the procedure outlined in Vergouwe et al (2016) and estimated four logistic regression models. The first included the HOMR linear predictor, with its coefficient set equal to 1, and intercept set to zero (the original HOMR model). The second model allowed the intercept to be freely estimated (Recalibration in the Large). The third then allowed the coefficient on the HOMR linear predictor to be freely estimated as well (Logistic Recalibration). Finally, the fourth model included the complete set variables used in the HOMR model, including the same transformations and interactions, and allowed their respective coefficients to be freely estimated (Model Revision). Here is the code for executing those models:
# HOMR (intercept = 0 and beta = 1)
m1 <- glm(alive ~ -1 + offset(z_homr), data = data, family = binomial)
# Recalibration in the Large (estimate intercept, beta = 1)
m2 <- glm(alive ~ 1 + offset(z_homr), data = data, family = binomial)
# Logisitic Calibration (estimate both)
m3 <- glm(alive ~ 1 + z_homr, data = data, family = binomial)
# Model Revision
data2 <- data
data2$admit_service <- as.character(data2$admit_service)
data2$admit_service <- factor(
ifelse( # Sparse cells, so this was collapsed to binary
data2$admit_service == "General Medicine",
data2$admit_service,
"Other"
)
)
formula_4 <- as.formula(
"alive ~ 1 + drs + sqrt_age + sex + living_status + log_cci +
trans_ed + trans_amb + admit_service + urgency_admit + urgent_readmit +
(sqrt_age * log_cci) + (living_status * trans_amb) +
(urgency_admit * trans_amb)"
)
m4 <- glm(formula_4, data = data2, family = binomial)
# Model predicted probabilties
data$m1_pred <- predict(m1, type = "response")
data$m2_pred <- predict(m2, type = "response")
data$m3_pred <- predict(m3, type = "response")
data$m4_pred <- predict(m4, type = "response")
The overall performance of these models was evaluated using the Brier score, rescaled to range from 0 to 1 (with higher values indicating better performance) as suggested by Steyerberg et al (2010). We assessed calibration graphically, in addition to using the maximum and average difference in predicted vs loess-calibrated probabilities (Emax and Eavg); and we used the c-statistic to assess discrimination. For each of these metrics, we reported bootstrapped 95% confidence intervals. Finally, for Model Revision, we estimated an optimism-corrected c-statistic and shrinkage factor using bootstrapping, as described in Harrell, Lee and Mark (1996)
To get most metrics, we used rms::val.prob as follows:
# Metrics
val_m1 <- val.prob(data$m1_pred, as.numeric(data$alive) - 1,
pl = FALSE) %>% round(3)
val_m2 <- val.prob(data$m2_pred, as.numeric(data$alive) - 1,
pl = FALSE) %>% round(3)
val_m3 <- val.prob(data$m3_pred, as.numeric(data$alive) - 1,
pl = FALSE) %>% round(3)
val_m4 <- val.prob(data$m4_pred, as.numeric(data$alive) - 1,
pl = FALSE) %>% round(3)
rescale_brier <- function(x, p, ...){
format(round(1 - (x / (mean(p) * (1 - mean(p)))), digits = 2), nsmall = 2)
}
b1 <- rescale_brier(val_m1["Brier"], 0.184)
b2 <- rescale_brier(val_m2["Brier"], 0.184)
b3 <- rescale_brier(val_m3["Brier"], 0.184)
b4 <- rescale_brier(val_m4["Brier"], 0.184)
# Note: 0.184 is the marginal probabilty of death in the entire sample
Here is what the rms::val.prob output looks like.
pander(val_m1)
Table continues below
| 0.564 |
0.782 |
0.22 |
0.145 |
204.6 |
NA |
0.021 |
32.23 |
0 |
| 0.123 |
0.127 |
-0.428 |
0.985 |
0.103 |
0.102 |
0.058 |
-3.806 |
0 |
Uncertainty in these metrics was evaluated with bootstrapping.
set.seed(48572)
boot_val <- function(data, formula, ...){
out <- list()
for(i in 1:500){
df <- sample_n(data, nrow(data), replace = TRUE)
md <- glm(formula, data = df, family = binomial)
out[[i]] <- val.prob(predict(md, type = "response"),
as.numeric(df$alive) - 1,
pl = FALSE) %>% round(3)
}
return(out)
}
boot_vals_m1 <- boot_val(data, as.formula("alive ~ -1 + offset(z_homr)"))
boot_vals_m2 <- boot_val(data, as.formula("alive ~ 1 + offset(z_homr)"))
boot_vals_m3 <- boot_val(data, as.formula("alive ~ 1 + z_homr "))
boot_vals_m4 <- boot_val(data2, formula_4)
This code just pulls out 95% intervals from the bootstrapped values.
calc_ci <- function(metric, boot_vals, n){
x <- unlist(map(boot_vals, `[`, c(metric)))
if(metric == 'Brier'){x <- as.numeric(rescale_brier(x, 0.184))}
paste0("(", round(quantile(x, 0.025), n), " to ",
round(quantile(x, 0.975), n), ")")
}
# m1
m1_c_boot_ci <- calc_ci("C (ROC)", boot_vals_m1, 2)
m1_brier_boot_ci <- calc_ci("Brier", boot_vals_m1, 2)
m1_emax_boot_ci <- calc_ci("Emax", boot_vals_m1, 2)
m1_eavg_boot_ci <- calc_ci("Eavg", boot_vals_m1, 2)
# m2
m2_c_boot_ci <- calc_ci("C (ROC)", boot_vals_m2, 2)
m2_brier_boot_ci <- calc_ci("Brier", boot_vals_m2, 2)
m2_emax_boot_ci <- calc_ci("Emax", boot_vals_m2, 2)
m2_eavg_boot_ci <- calc_ci("Eavg", boot_vals_m2, 2)
# m3
m3_c_boot_ci <- calc_ci("C (ROC)", boot_vals_m3, 2)
m3_brier_boot_ci <- calc_ci("Brier", boot_vals_m3, 2)
m3_emax_boot_ci <- calc_ci("Emax", boot_vals_m3, 2)
m3_eavg_boot_ci <- calc_ci("Eavg", boot_vals_m3, 2)
# m4
m4_c_boot_ci <- calc_ci("C (ROC)", boot_vals_m4, 2)
m4_brier_boot_ci <- calc_ci("Brier", boot_vals_m4, 2)
m4_emax_boot_ci <- calc_ci("Emax", boot_vals_m4, 2)
m4_eavg_boot_ci <- calc_ci("Eavg", boot_vals_m4, 2)
This code is for the shrinkage corrected c-index and calibration slope for Model Revision. Since we are estimating new coefficients for each predictor from the data we have, this helps us avoid over-fit. To do this, we use rms::validate.
# To use validate, we need to estimate m4 with lrm instead of glm.
d <- datadist(data)
options(datadist = "d")
m4b <- lrm(formula_4, data = data2, x = TRUE, y = TRUE)
set.seed(04012019)
val_new <- rms::validate(m4b, B = 500)
shrink_factor <- round(val_new["Slope","index.corrected"], 2)
c_corrected <- round(0.5 * (1 + val_new["Dxy","index.corrected"]), 2)
Finally, this is the code for making the calibration plots (rms::val.plot will also give a nice calibration plot unless it’s suppressed in the call, but I wanted a ggplot based version so I could tweak it to my liking).
# Function to produce the calibration plots
cal_plot <- function(model, model_name, pred_var, ...){
require(tidyverse)
require(viridis)
require(gridExtra)
# The calibration plot
g1 <- mutate(data, bin = ntile(get(pred_var), 10)) %>%
# Bin prediction into 10ths
group_by(bin) %>%
mutate(n = n(), # Get ests and CIs
bin_pred = mean(get(pred_var)),
bin_prob = mean(as.numeric(alive) - 1),
se = sqrt((bin_prob * (1 - bin_prob)) / n),
ul = bin_prob + 1.96 * se,
ll = bin_prob - 1.96 * se) %>%
ungroup() %>%
ggplot(aes(x = bin_pred, y = bin_prob, ymin = ll, ymax = ul)) +
geom_pointrange(size = 0.5, color = "black") +
scale_y_continuous(limits = c(0, 1), breaks = seq(0, 1, by = 0.1)) +
scale_x_continuous(limits = c(0, 1), breaks = seq(0, 1, by = 0.1)) +
geom_abline() + # 45 degree line indicating perfect calibration
geom_smooth(method = "lm", se = FALSE, linetype = "dashed",
color = "black", formula = y~-1 + x) +
# straight line fit through estimates
geom_smooth(aes(x = get(pred_var), y = as.numeric(alive) - 1),
color = "red", se = FALSE, method = "loess") +
# loess fit through estimates
xlab("") +
ylab("Observed Probability") +
theme_minimal() +
ggtitle(model_name)
# The distribution plot
g2 <- ggplot(data, aes(x = get(pred_var))) +
geom_histogram(fill = "black", bins = 200) +
scale_x_continuous(limits = c(0, 1), breaks = seq(0, 1, by = 0.1)) +
xlab("Predicted Probability") +
ylab("") +
theme_minimal() +
scale_y_continuous(breaks = c(0, 40)) +
theme(panel.grid.minor = element_blank())
# Combine them
g <- arrangeGrob(g1, g2, respect = TRUE, heights = c(1, 0.25), ncol = 1)
grid.newpage()
grid.draw(g)
return(g[[3]])
}
Results
# Combine all the results into a single dataframe
model_tab_1 <- data_frame(
est = c("Intercept", "Slope", "Residual deviance", "Df",
"LRT Chisq p-value", "Brier score (rescaled)",
"Emax", "Eavg", "c-statistic"),
m1_est = c(round(c(0, 1, m1$deviance, m1$df.residual, 0), 2),
paste(b1, m1_brier_boot_ci),
paste(val_m1["Emax"], m1_emax_boot_ci),
paste(val_m1["Eavg"], m1_eavg_boot_ci),
paste(round(val_m1["C (ROC)"], 2), m1_c_boot_ci)),
m2_est = c(round(c(tidy(m2)$estimate, 1, m2$deviance, m2$df.residual, 0), 2),
paste(b2, m2_brier_boot_ci),
paste(val_m2["Emax"], m2_emax_boot_ci),
paste(val_m2["Eavg"], m2_eavg_boot_ci),
paste(round(val_m2["C (ROC)"], 2), m2_c_boot_ci)),
m3_est = c(round(c(tidy(m3)$estimate, m3$deviance, m3$df.residual, 0), 2),
paste(b3, m3_brier_boot_ci),
paste(val_m3["Emax"], m3_emax_boot_ci),
paste(val_m3["Eavg"], m3_eavg_boot_ci),
paste(round(val_m3["C (ROC)"], 2), m3_c_boot_ci)),
m4_est = c("", "", round(c(m4$deviance, m4$df.residual, 0), 2),
paste(b4, m4_brier_boot_ci),
paste(val_m4["Emax"], m4_emax_boot_ci),
paste(val_m4["Eavg"], m4_eavg_boot_ci),
paste(round(val_m4["C (ROC)"], 2), m4_c_boot_ci))
)
names(model_tab_1) <- c("", "HOMR model", "Calibration in the Large",
"Logistic Recalibration", "Model Revision")
model_tab_1[5, 2:5] <- c(
"-", "<0.001", round(anova(m2, m3, test = "Chisq")[5][2, ], 2), "-"
)
Table 1. All the results hacked into a table.
knitr::kable(model_tab_1)
| Intercept |
0 |
-0.42 |
-0.43 |
|
| Slope |
1 |
1 |
0.99 |
|
| Residual deviance |
1139.96 |
1107.76 |
1107.73 |
1046.55 |
| Df |
1409 |
1408 |
1407 |
1389 |
| LRT Chisq p-value |
- |
<0.001 |
0.85 |
- |
| Brier score (rescaled) |
0.15 (0.09 to 0.22) |
0.19 (0.11 to 0.26) |
0.19 (0.11 to 0.27) |
0.23 (0.17 to 0.32) |
| Emax |
0.103 (0.08 to 0.17) |
0.111 (0.03 to 0.25) |
0.121 (0.03 to 0.23) |
0.017 (0.01 to 0.11) |
| Eavg |
0.058 (0.04 to 0.08) |
0.016 (0.01 to 0.03) |
0.017 (0.01 to 0.03) |
0.008 (0 to 0.02) |
| c-statistic |
0.78 (0.75 to 0.81) |
0.78 (0.75 to 0.81) |
0.78 (0.75 to 0.81) |
0.82 (0.8 to 0.86) |
Calibration plots
Figure 2. Calibration plot for the HOMR model in a sample of 1409 patients aged 65 years or older that were under the care of geriatric medicine service at Cork University Hospital (2013-01-01 to 2015-03-06)
x <- cal_plot(m1, "HOMR model", "m1_pred")

Figure 3. Calibration plot for Recalibration in the Large 
Figure 4. Calibration plot for Model Revision 
Other stuff
Figure 5. Logistic curve fit of the original HOMR model to one year post-hospitalization mortality
# Plot of data with a logistic curve fit
ggplot(data, aes(x = z_homr, y = as.numeric(alive) - 1)) +
geom_jitter(height = 0.1, size =1, alpha = 0.5) +
geom_smooth(method = "glm",
method.args = list(family = "binomial")) +
theme_minimal() +
scale_y_continuous(breaks = c(0, 1), labels = c("Alive", "Dead")) +
ylab("") +
xlab("HOMR Linear Predictor")

Figure 6. Number/Proportion of patients who died within one year of hospitalization by risk level (Z)
g1 <- ggplot(data, aes(x = z_homr, fill = alive)) +
geom_histogram() +
theme_minimal() +
xlab("HOMR Linear Predictor") +
ylab("Number of Participants") +
scale_fill_brewer("Alive", palette = "Paired")
g2 <- ggplot(data, aes(x = z_homr, fill = alive)) +
geom_histogram(position = "fill") +
theme_minimal() +
xlab("HOMR Linear Predictor") +
ylab("Proportion") +
scale_fill_brewer("Alive", palette = "Paired")
grid.arrange(g1, g2, ncol = 1)

Figure 7. Distribution of the original HOMR linear predictor among those who did and didn’t die within one year after hospitalization
ggplot(data, aes(y = z_homr, x = alive, fill = alive, color = alive)) +
geom_beeswarm() +
geom_boxplot(alpha = 0, color = "black") +
theme_minimal() +
ylab("HOMR Linear Predictor") +
xlab("Alive at 1 year") +
scale_fill_brewer(guide = FALSE, palette = "Paired") +
scale_color_brewer(guide = FALSE, palette = "Paired")

Table 2. HOMR Model Revision coefficients
covariates <- c(
"Intercept",
"DRS",
"sqrt(Age)",
"Male (vs Female)",
"Rehab",
"Homecare",
"Nursing Home",
"log(CCI)",
"sqrt(Ed visits in the previous year + 1)",
"1/(Admissions by ambulance in previous year +1)",
"Other (vs General Medicine)",
"ED w/o Ambulance",
"ED w/Ambulance",
"Urgent readmission",
"Sqrt(Age) * log(CCI)",
"Rehab * 1/(Admissions by ambulance in previous year +1)",
"Homecare * 1/(Admissions by ambulance in previous year +1)",
"Nursing Home * 1/(Admissions by ambulance in previous year +1)",
"ED w/o Ambulance * 1/(Admissions by ambulance in previous year +1)",
"ED w/Ambulance * 1/(Admissions by ambulance in previous year +1)"
)
sjPlot::tab_model(m4, show.p = FALSE, show.ci = FALSE, show.se = TRUE,
transform = NULL, pred.labels = covariates)
|
|
alive
|
|
Predictors
|
Log-Odds
|
std. Error
|
|
Intercept
|
-14.84
|
4.02
|
|
DRS
|
0.11
|
0.02
|
|
sqrt(Age)
|
1.45
|
0.43
|
|
Male (vs Female)
|
0.44
|
0.17
|
|
Rehab
|
-1.16
|
0.71
|
|
Homecare
|
0.41
|
0.66
|
|
Nursing Home
|
-0.34
|
1.29
|
|
log(CCI)
|
2.78
|
2.83
|
|
sqrt(Ed visits in the previous year + 1)
|
0.16
|
0.71
|
|
1/(Admissions by ambulance in previous year +1)
|
0.19
|
0.61
|
|
Other (vs General Medicine)
|
-0.68
|
0.46
|
|
ED w/o Ambulance
|
0.38
|
0.67
|
|
ED w/Ambulance
|
1.21
|
1.12
|
|
Urgent readmission
|
0.60
|
0.27
|
|
Sqrt(Age) * log(CCI)
|
-0.23
|
0.31
|
|
Rehab * 1/(Admissions by ambulance in previous year +1)
|
-0.31
|
0.79
|
|
Homecare * 1/(Admissions by ambulance in previous year +1)
|
-0.51
|
0.82
|
|
Nursing Home * 1/(Admissions by ambulance in previous year +1)
|
-0.46
|
1.78
|
|
ED w/o Ambulance * 1/(Admissions by ambulance in previous year +1)
|
-0.87
|
0.76
|
|
ED w/Ambulance * 1/(Admissions by ambulance in previous year +1)
|
-1.91
|
1.34
|
|
Observations
|
1409
|
|
Cox & Snell’s R2 / Nagelkerke’s R2
|
0.191 / 0.310
|
Note: Admit service recoded to General Medicine vs Other, due to small call sizes. ICU admission was omitted as there were only 3 cases of this happening. Home O2 was omitted since no patients in our sample were using it.
Full distributions of bootstrapped values.
boots <- function(metric, boot_vals){
x <- as.numeric(unlist(map(boot_vals, `[`, metric)))
if(metric == 'Brier'){x <- as.numeric(rescale_brier(x, 0.184))}
return(x)
}
boot_list <- list(boot_vals_m1, boot_vals_m2, boot_vals_m3, boot_vals_m4)
metrics <- c("C (ROC)", "Brier", "Eavg", "Emax")
x <- c()
for(i in metrics){
for(j in 1:4){
x <- c(x, boots(i, boot_list[[j]]))
}
}
y <- rep(c(paste0("m", 1:4, " c-index"), paste0("m", 1:4, " Brier"),
paste0("m", 1:4, " Emax"), paste0("m", 1:4, " Eavg")),
each = 500)
boot_data <- data_frame(var = y, val = x)
Figure 7. Distributions of bootstrapped model statistics
ggplot(boot_data, aes(x = val)) +
geom_density() +
facet_wrap(~var, nrow = 4, scales = "free") +
theme_minimal() +
xlab("") +
ylab("Density")

# Packages used
# library(tidyverse)
# library(rms)
# library(Hmisc)
# library(knitr)
# library(broom)
# library(pander)
# library(ggbeeswarm)
# library(gridExtra)
# library(grid)
# library(sjPlot)
# library(sjmisc)
# library(sjlabelled)
# library(viridis)
sessionInfo()
## R version 3.4.3 (2017-11-30)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 15063)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_United Kingdom.1252
## [2] LC_CTYPE=English_United Kingdom.1252
## [3] LC_MONETARY=English_United Kingdom.1252
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United Kingdom.1252
##
## attached base packages:
## [1] grid methods stats graphics grDevices utils datasets
## [8] base
##
## other attached packages:
## [1] viridis_0.5.1 viridisLite_0.3.0 sjlabelled_1.0.17
## [4] sjmisc_2.7.9 sjPlot_2.6.2 gridExtra_2.3
## [7] ggbeeswarm_0.6.0 pander_0.6.2 broom_0.4.4
## [10] knitr_1.20 rms_5.1-2 SparseM_1.77
## [13] Hmisc_4.1-1 Formula_1.2-3 survival_2.41-3
## [16] lattice_0.20-35 forcats_0.3.0 stringr_1.3.1
## [19] dplyr_0.8.0.1 purrr_0.2.5 readr_1.1.1
## [22] tidyr_0.8.1 tibble_2.0.1 ggplot2_3.1.0
## [25] tidyverse_1.2.1
##
## loaded via a namespace (and not attached):
## [1] nlme_3.1-131 lubridate_1.7.4 insight_0.2.0
## [4] RColorBrewer_1.1-2 httr_1.4.0 rprojroot_1.3-2
## [7] TMB_1.7.13 tools_3.4.3 backports_1.1.2
## [10] R6_2.2.2 rpart_4.1-11 vipor_0.4.5
## [13] lazyeval_0.2.1 colorspace_1.3-2 nnet_7.3-12
## [16] withr_2.1.2 tidyselect_0.2.5 mnormt_1.5-5
## [19] emmeans_1.2.3 curl_3.3 compiler_3.4.3
## [22] cli_1.0.1 rvest_0.3.2 quantreg_5.36
## [25] htmlTable_1.12 xml2_1.2.0 sandwich_2.4-0
## [28] labeling_0.3 bookdown_0.9 scales_1.0.0
## [31] checkmate_1.8.5 polspline_1.1.13 mvtnorm_1.0-8
## [34] psych_1.8.4 digest_0.6.18 minqa_1.2.4
## [37] foreign_0.8-69 rmarkdown_1.10 base64enc_0.1-3
## [40] pkgconfig_2.0.2 htmltools_0.3.6 lme4_1.1-17
## [43] highr_0.7 htmlwidgets_1.3 rlang_0.3.4
## [46] readxl_1.1.0 rstudioapi_0.7 zoo_1.8-2
## [49] jsonlite_1.6 acepack_1.4.1 magrittr_1.5
## [52] Matrix_1.2-12 Rcpp_1.0.0 munsell_0.5.0
## [55] stringi_1.4.3 multcomp_1.4-8 yaml_2.1.19
## [58] snakecase_0.9.1 MASS_7.3-47 plyr_1.8.4
## [61] parallel_3.4.3 crayon_1.3.4 ggeffects_0.9.0
## [64] haven_2.1.0 splines_3.4.3 sjstats_0.17.4
## [67] hms_0.4.2 pillar_1.3.1 estimability_1.3
## [70] reshape2_1.4.3 codetools_0.2-15 glue_1.3.0
## [73] evaluate_0.10.1 blogdown_0.6 latticeExtra_0.6-28
## [76] data.table_1.11.4 modelr_0.1.2 nloptr_1.0.4
## [79] MatrixModels_0.4-1 cellranger_1.1.0 gtable_0.2.0
## [82] assertthat_0.2.0 xfun_0.2 xtable_1.8-2
## [85] coda_0.19-1 glmmTMB_0.2.1.0 beeswarm_0.2.3
## [88] cluster_2.0.6 TH.data_1.0-8